Regulation of Gene Expression Study Guide
Introduction
Living systems store, retrieve, transmit and respond to information essential to life processes. Expression of genetic information involves cellular and molecular mechanisms. Gene regulation results in differential gene expression, leading to cell specialization.
Prokaryotic gene expression
The fundamental method to control the group of proteins and how much of each protein is expressed in a prokaryotic cell is the regulation of DNA transcription. Hence, in prokaryotic cells, the control of gene expression is mostly at the transcriptional level.
- Proteins needed for a specific function or involved in the same biochemical pathway are encoded together in blocks called operons.
- Each operon contains regulatory DNA sequences, that act as binding sites for regulatory proteins which promote or inhibit transcription.
- Regulatory proteins bind to tiny molecules, which can make the protein active or inactive by changing its ability to bind DNA.
There are 3 types of regulatory molecules that can affect the expression of operons in prokaryotic cells: repressors, activators, and inducers.
- Both activators and repressors regulate gene expression by binding to specific DNA sites adjacent to the genes they control.
- Generally, activators bind to the promoter site and repressors bind to operator regions.
- Repressors inhibit transcription of a gene in response to an external stimulus while activators increase the transcription of a gene in response to an external stimulus.
- Inducers are tiny molecules that could be produced by the cell or present in the cell’s environment. Based on the needs of the cell and the availability of substrate, inducers can either activate or repress transcription.
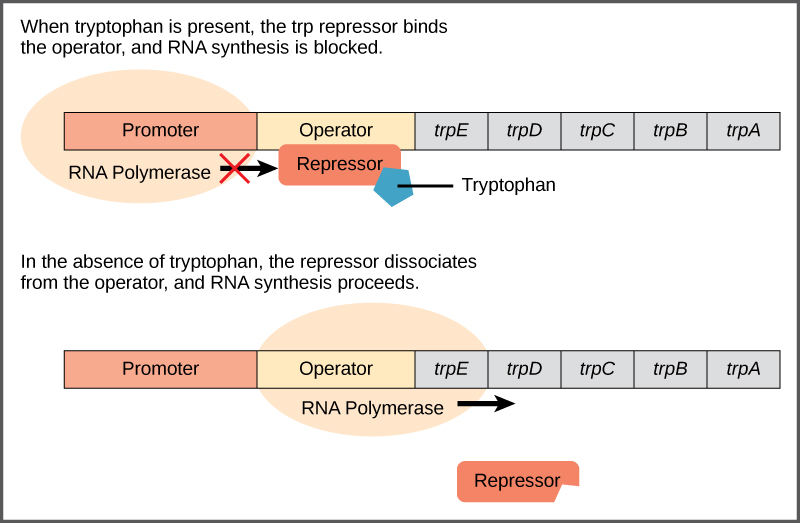
The trp Operon: A Repressible Operon
- The trp operon, found in E. coli bacteria, is a group of genes that encode biosynthetic enzymes for the amino acid tryptophan.
- The trp operon is expressed (“on”) when tryptophan levels are low and repressed (“off”) when the levels are high.
- The trp repressor regulates the trp operon. When bound to tryptophan, the trp repressor blocks the expression of the operon.
- Tryptophan biosynthesis is also regulated by attenuation (a mechanism based on the coupling of transcription and translation).
In the tryptophan operon, the 5 genes that are needed to synthesize tryptophan in E. coli are present next to each other.
- When tryptophan is abundant, two tryptophan molecules bind the repressor protein at the operator sequence. This helps block the RNA polymerase from transcribing the tryptophan genes.
- When tryptophan is absent, the repressor protein would not bind to the operator, and the genes are transcribed.
- As the repressor protein actively binds to the operator to keep the genes turned off, the trp operon is considered to be negatively regulated, and the proteins that bind to the operator to silence trp expression are negative regulators.
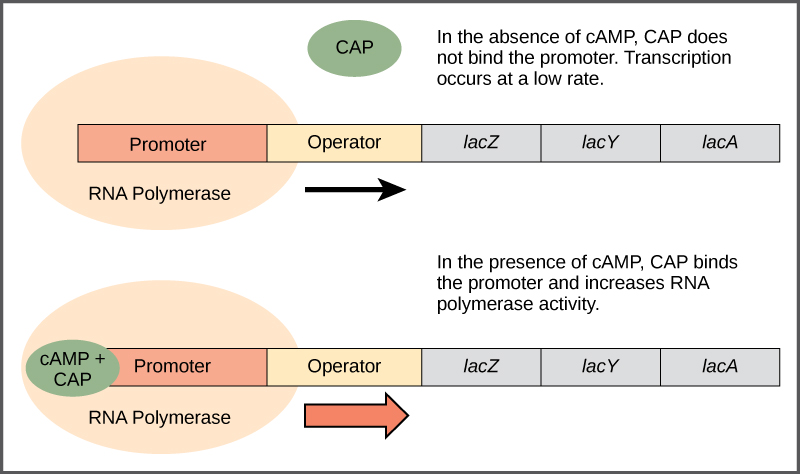
Catabolite Activator Protein (CAP): A Transcriptional Activator
- Catabolite activator protein (CAP) acts as a glucose sensor. When glucose levels are low, it activates transcription of the operon. CAP can sense glucose indirectly through the “hunger signal” molecule cAMP.
- The CAP binding site is a positive regulatory site that is bound by catabolite activator protein (CAP). When CAP is bound to this site, it promotes transcription by helping RNA polymerase bind to the promoter.
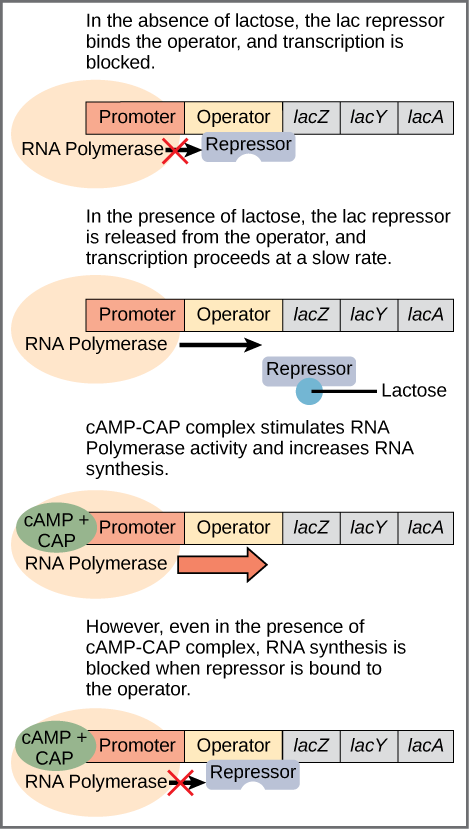
The lac Operon: An Inducible Operon
- The lac operon of E. coli has genes involved in lactose metabolism, which is expressed only when lactose is present, and glucose is absent.
- The regulators, lac repressor and catabolite activator protein (CAP), turn the operon “on” and “off” in response to lactose and glucose levels.
- The lac repressor acts as a lactose sensor and it normally blocks transcription of the operon but stops acting as a repressor under the presence of lactose. The lac repressor possess the capability to sense lactose indirectly through its isomer allolactose.
- The promoter is the binding site for RNA polymerase (the enzyme that performs transcription).
- The operator is a negative regulatory site bound by the lac repressor protein. The operator overlaps with the promoter, and when the lac repressor is bound, RNA polymerase cannot bind to the promoter and start transcription.
The lac operon would be expressed at high levels if both the following conditions are met:
-
Glucose must be unavailable: When glucose is unavailable, cAMP binds to CAP, making CAP able to bind DNA. Bound CAP helps RNA polymerase attach to the lac operon promoter.
-
Lactose must be available: If lactose is available, the lac repressor will be released from the operator (by binding of allolactose). This allows RNA polymerase to move forward on the DNA and transcribe the operon.
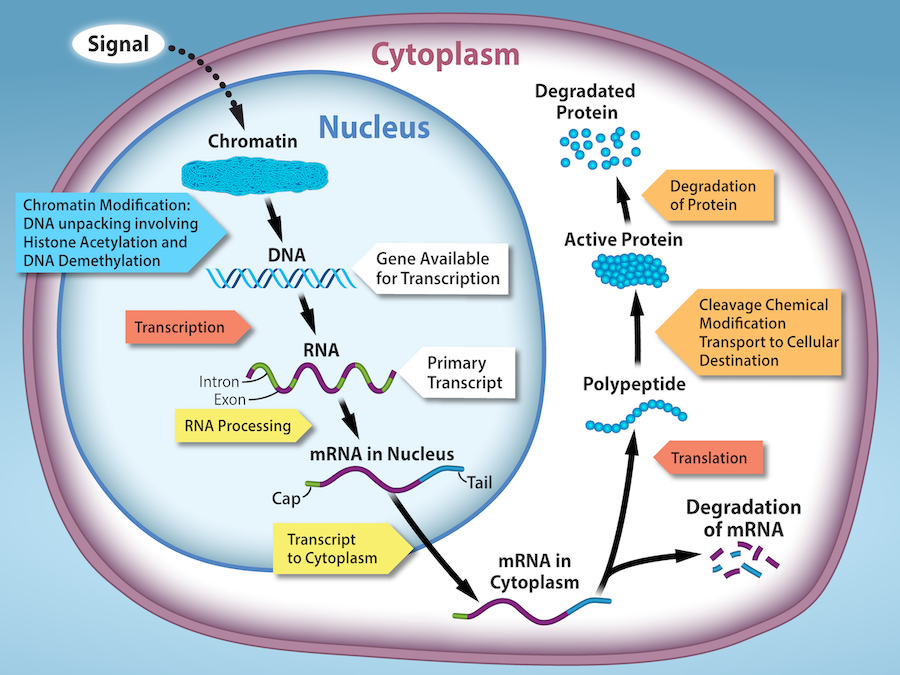
Eukaryotic gene regulation
In eukaryotes, gene expression is regulated during transcription and RNA processing, which takes place in the nucleus during protein translation. The regulation of gene expression in this case can occur at all stages of the process.
Regulation can occur :
- When the DNA is uncoiled and loosened from nucleosomes to bind transcription factors (epigenetic level) or
- When the RNA is transcribed (transcriptional level) or
- When the RNA is processed and exported to the cytoplasm after it has been transcribed (post-transcriptional level) or
- When the RNA is translated into protein (translational level), or
- After the protein is made (post-translational level).
Although eukaryotic gene expression involves many steps, almost all of them can be regulated. Various genes are regulated at various points, and it’s not uncommon for a gene to be regulated at multiple steps.
- Chromatin accessibility. Regulation of the structure of chromatin (DNA and its organizing proteins) can occur. More open chromatin would make a gene better available for transcription.
- Transcription. Transcription is a primary regulatory point for many genes. Sets of transcription factor proteins can bind to certain DNA sequences in or near a gene to promote or repress its transcription into an RNA.
- RNA processing. Several mRNAs can be made from the same pre-mRNA by alternative splicing.
- RNA stability. The number of proteins that can be made from an mRNA molecule depends on the lifetime of the mRNA molecule in the cytosol. Small regulatory RNAs called miRNAs can bind to target mRNAs and divide them.
- Translation. Regulators can increase or inhibit the translation of an mRNA.
- Protein activity. Proteins undergo a variety of modifications and these modifications can be regulated. This could affect the activity or behavior of the protein.
Conclusion:
- The process of turning on a gene to produce RNA and protein is called gene expression.
- Whether in a simple unicellular organism or a complex multi-cellular organism, each cell controls when and how its genes are expressed.
- In prokaryotic cells, the control of gene expression is mostly at the transcriptional level.
- In eukaryotic cells, the regulation of gene expression can occur at all stages of the process. Regulation may occur when the DNA is uncoiled and loosened from nucleosomes to bind transcription factors (epigenetic level), when the RNA is transcribed (transcriptional level), when the RNA is processed and exported to the cytoplasm after it is transcribed (post-transcriptional level), when the RNA is translated into protein (translational level), or after the protein has been made (post-translational level).
FAQs
1. Differentiate between negative versus positive regulation.
In positive regulation, genes undergo transcription and the binding of specific protein (activator) is required for transcription to begin.In negative regulation, the binding of a specific protein (repressor) inhibits transcription from occurring.
2. What stage of gene expression is the primary point for specific gene regulation?
Although, gene regulation can occur at any point of the transcription-translation process, the primary point is at the transcription level.
3. Why is regulation of gene expression important?
The regulation of gene expression conserves energy and space. It would require a significant amount of energy for an organism to express every gene at all times, so it is more energy efficient to turn on the genes only when they are required.
4. Which form of vitamin A is involved in the regulation of gene expression?
All-trans-Retinoic acid (RA) is the major biologically active form of vitamin A involved in the regulation of gene expression.
5. Explain why cap binding and stimulation of gene expression is positive regulation.
CAP binding with the cAMP increases the rate of transcription (gene expression). It is a positive regulation because it is continuously increasing.
We hope you enjoyed studying this lesson and learned something cool about Regulation of Gene Expression! Join our Discord community to get any questions you may have answered and to engage with other students just like you! Don’t forget to download our App to experience our fun VR classrooms – we promise, it makes studying much more fun 😎
Sources:
- “16.2 Prokaryotic Gene Regulation – Biology for AP® Courses | OpenStax.” Openstax.org, openstax.org/books/biology-ap-courses/pages/16-2-prokaryotic-gene-regulation. Accessed 12 Feb. 2022.
- “Eukaryotic Epigenetic Gene Regulation – Biology for AP® Courses – OpenStax.” Openstax.org, openstax.org/books/biology-ap-courses/pages/16-3-eukaryotic-epigenetic-gene-regulation. Accessed 12 Feb. 2022.
- “Regulation of Gene Expression | Biology for Majors I.” Courses.lumenlearning.com, courses.lumenlearning.com/suny-wmopen-biology1/chapter/regulation-of-gene-expression/#:~:text=The%20regulation%20of%20gene%20expression. Accessed 12 Feb. 2022.